Single-molecule level DNA-protein interaction studies
DNA-protein interactions play a key role in many biological processes. One unifying aspect of these interactions is target search process. It involves different mechanisms that are different for each protein, but can be separated into these classes: 3D diffusion (i.e., jumping), sliding, and hopping. Those interactions can differ in number of DNA target sites that protein can bind. DNA-protein interactions can occur, when one protein interacts with two targets either on a single DNA molecule (i.e. in Cis interaction) or on two separate molecules (i.e. in Trans interaction).
DNA-protein interaction occurs on a nanometer length scale at millisecond-to-second timescales. To monitor interactions occurring on the seconds time scale surface immobilization of one of interacting partners is often required. These interactions can be visualized at great detail through: force spectroscopy, fluorescence, Forster resonance energy transfer (FRET), atomic force microscopy (AFM), nanopores, DNA flow-stretch assays, etc [A. Kopūstas, et al. Applied Nano, 2022]. Single-molecule FRET (smFRET) approach worked very well for many different protein-DNA interaction studies.
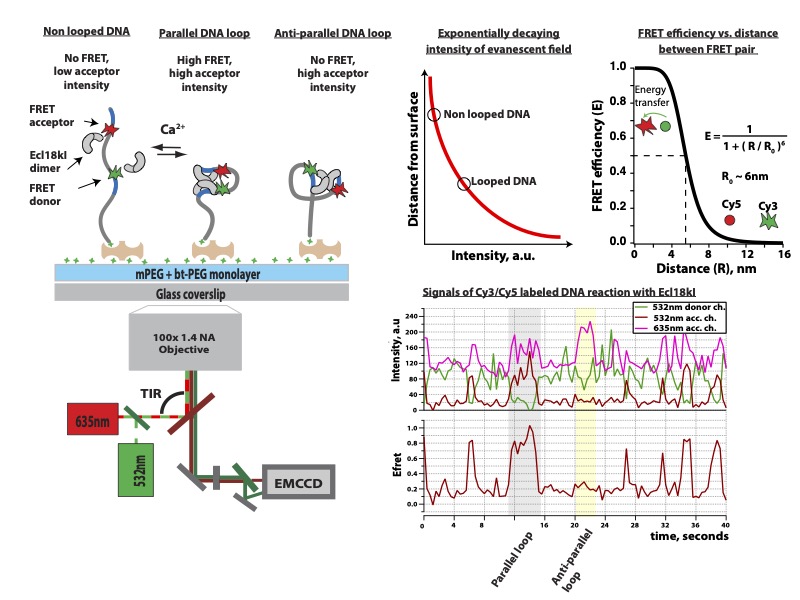
Restriction endonucleases (REases) in bacteria and archaea function as defense against invading foreign DNA systems. Over the past 10 years we studied different REases using smFRET approach: Ecl18kI protein [M. Tutkus, et al. Biophysical J. 2017] , and NgoMIV [M. Tutkus, et al. Biopolymers. 2017]. Since both proteins recognize a palindromic DNA target sequence, DNA fragment in our DNA looping assay could adopt two type of loops (“U”- or “Phi”-shaped) depending on how each copy of protein was oriented on the target site (see figure below this paragraph). In our assay we monitored events of DNA looping that occurred after proteins’ binding onto target sites, and events of DNA loop release that occurred upon proteins dissociation from target sites.
Recently our team developed an alternative assay to the original DNA Curtains that we termed the Soft DNA Curtains. We fabricated streptavidin or traptavidin patterns (i.e. line-features) on the modified coverslip surface that can be utilized to assemble stably immobilized biotinylated DNA arrays. The application of hydrodynamic buffer flow allows extension of the immobilized DNA molecules along the surface of the flowcell channel. This year we published a follow-up article in Langmuir on the Oriented Soft DNA Curtains. In this work, we fabricated the uniformly oriented double-tethered DNA Curtains using heterologous labeling of the DNA by biotin and digoxigenin. In addition to that we increased stability of the immobilized DNA molecules using a more stable alternative to sAv called traptavidin as an ink for the fabrication of protein templates. The main goal of our research is to apply the developed platform for studies of DNA targeting mechanisms of diverse CRISPR-Cas (Clustered regulatory interspaced short palindromic repeats) systems family, novel molecular-tools – prokaryotic Argonaute (pAgo) proteins, and various restriction endonucleases. Since 2017 for these studies we received two grants from the Lithuanian research council. Also, we received funding for a post-doc project dedicated for pAgo studies. 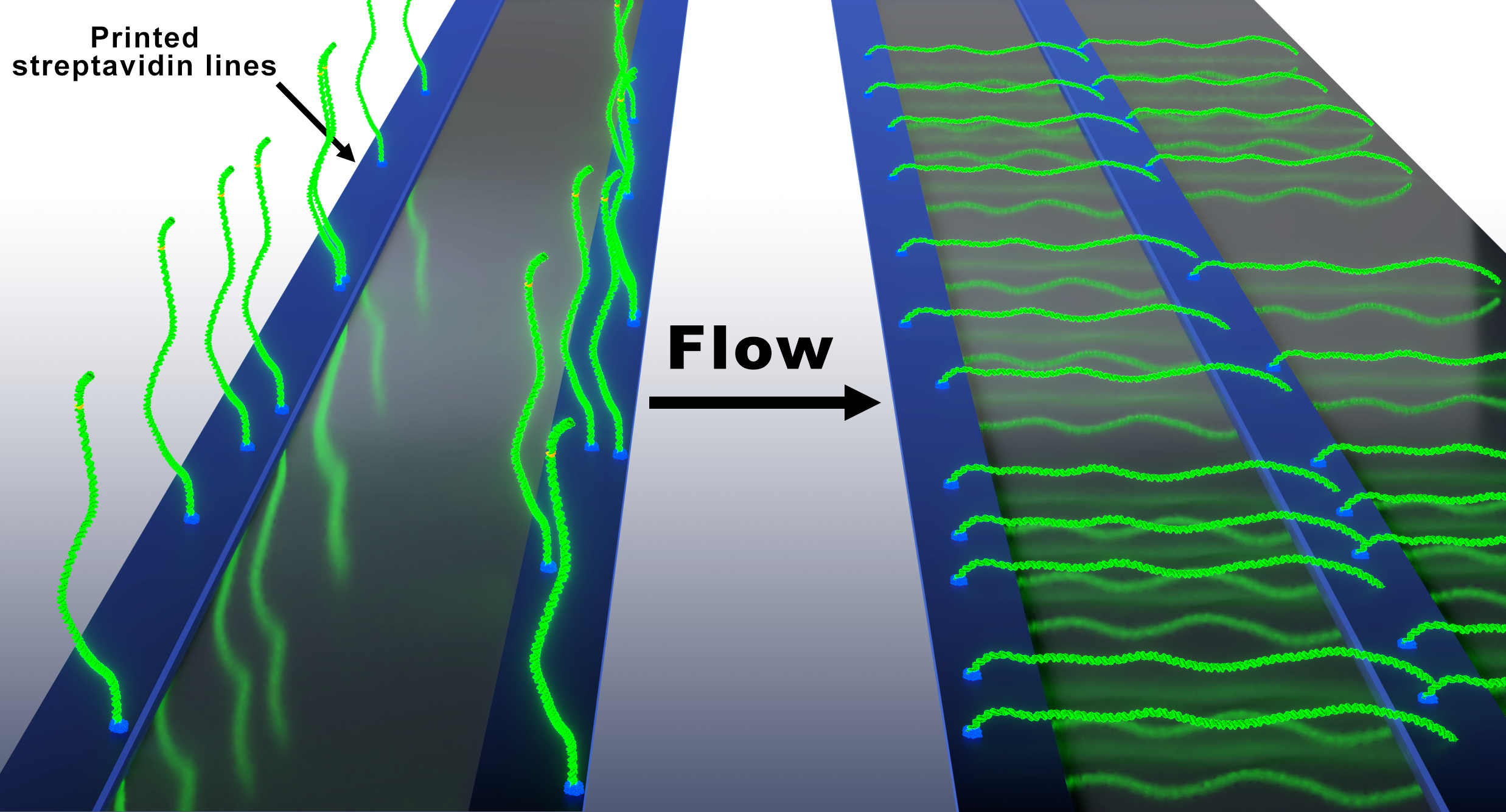
our developed Igor Pro software for fluorescent spot intensity extraction and analysis
MT Team 2022
MT Team 2023 September
Ph.D. Student - Vilnius University
Ph.D. Student - Vilnius University
MT Team 2024
MT Team 2025
MT Team during SMLMS2025
MSc. Student - Vilnius University
BSc. Neurobiophyics degree - Vilnius University
MSc. Neurobiophyics degree - Vilnius University (now Teacher)
MSc. Student - Vilnius University
MSc. Molecular Biology degree - Vilnius University
MSc. Student - Vilnius University
MSc. Student - Vilnius University
BSc. Student - Vilnius University
BSc. Student - Vilnius University
DNA-protein interaction occurs on a nanometer length scale at millisecond-to-second timescales. To monitor interactions occurring on the seconds time scale surface immobilization of one of interacting partners is often required. These interactions can be visualized at great detail through: force spectroscopy, fluorescence, Forster resonance energy transfer (FRET), atomic force microscopy (AFM), nanopores, DNA flow-stretch assays, etc [A. Kopūstas, et al. Applied Nano, 2022]. Single-molecule FRET (smFRET) approach worked very well for many different protein-DNA interaction studies.
Restriction endonucleases (REases) in bacteria and archaea function as defense against invading foreign DNA systems. Over the past 10 years we studied different REases using smFRET approach: Ecl18kI protein [M. Tutkus, et al. Biophysical J. 2017] , and NgoMIV [M. Tutkus, et al. Biopolymers. 2017]. Since both proteins recognize a palindromic DNA target sequence, DNA fragment in our DNA looping assay could adopt two type of loops (“U”- or “Phi”-shaped) depending on how each copy of protein was oriented on the target site (see figure below this paragraph). In our assay we monitored events of DNA looping that occurred after proteins’ binding onto target sites, and events of DNA loop release that occurred upon proteins dissociation from target sites.

Recently our team developed an alternative assay to the original DNA Curtains that we termed the Soft DNA Curtains. We fabricated streptavidin or traptavidin patterns (i.e. line-features) on the modified coverslip surface that can be utilized to assemble stably immobilized biotinylated DNA arrays. The application of hydrodynamic buffer flow allows extension of the immobilized DNA molecules along the surface of the flowcell channel. This year we published a follow-up article in Langmuir on the Oriented Soft DNA Curtains. In this work, we fabricated the uniformly oriented double-tethered DNA Curtains using heterologous labeling of the DNA by biotin and digoxigenin. In addition to that we increased stability of the immobilized DNA molecules using a more stable alternative to sAv called traptavidin as an ink for the fabrication of protein templates. The main goal of our research is to apply the developed platform for studies of DNA targeting mechanisms of diverse CRISPR-Cas (Clustered regulatory interspaced short palindromic repeats) systems family, novel molecular-tools – prokaryotic Argonaute (pAgo) proteins, and various restriction endonucleases. Since 2017 for these studies we received two grants from the Lithuanian research council. Also, we received funding for a post-doc project dedicated for pAgo studies.

our developed Igor Pro software for fluorescent spot intensity extraction and analysis
Team members:
MT Team
Ayush Ganguli
Aurimas Kopūstas
MT Team
MT Team
MT Team
Ugnė Bagdonaitė
Oskaras Venckus
Šarūnė Ivanovaitė
Justė Paksaitė
Simonas Ašmontas
Meda Jurevičiūtė
Monika Roliūtė
Aina El Ravvas
Guoda Taraškevičiūtė
Publications:
 | Liquid-liquid phase separation of alpha-synuclein increases the structural variability of fibrils formed during amyloid aggregation Mantas Ziaunys, Darius Sulskis, Dominykas Veiveris, *Aurimas Kopūstas*, Ruta Snieckute, Kamile Mikalauskaite, Andrius Sakalauskas, *Marijonas Tutkus*, Vytautas Smirnovas Published in The FEBS Journal, August 2024 (see publication ) - Research paper, co-author |
 | Formation of Calprotectin Inhibits Amyloid Aggregation of S100A8 and S100A9 Proteins Ieva Baronaitė, Darius Šulskis, Aurimas Kopu̅stas, *Marijonas Tutkus*, and Vytautas Smirnovas Published in ACS Chemical Neuroscience, April 2024 (see publication ) - Research paper, co-author |
 | smFRET Detection of Cis and Trans DNA Interactions by the BfiI Restriction Endonuclease *Šarūnė Ivanovaitė*, *Justė Paksaitė*, *Aurimas Kopūstas*, Giedrė Karzaitė, Danielis Rutkauskas, Arunas Silanskas, Giedrius Sasnauskas, Mindaugas Zaremba, Stephen K. Jones Jr., *and Marijonas Tutkus* Published in J. Phys. Chem. B, July 2023 (see publication ) - Research paper, corresponding author |
 | Advancing Understanding of DNA-BfiI Restriction Endonuclease Cis and Trans Interactions through smFRET Technology *Šarūnė Ivanovaitė*, *Justė Paksaitė*, *Aurimas Kopūstas*, Giedrė Karzaitė, Danielis Rutkauskas, Arunas Silanskas, Giedrius Sasnauskas, Mindaugas Zaremba, Stephen K. Jones Jr., *Marijonas Tutkus* Published in bioRxiv, April 2023 (see publication ) - Preprint, Corresponding author |
 | Disarming of type IF CRISPR-Cas surveillance complex by anti-CRISPR proteins AcrIF6 and AcrIF9 Egle Kupcinskaite, *Marijonas Tutkus*, *Aurimas Kopūstas*, *Simonas Ašmontas*, Marija Jankunec, Mindaugas Zaremba, Giedre Tamulaitiene, Tomas Sinkunas Published in Scientific reports, September 2022 (see publication ) - Research paper, co-author |
 | DNA Flow-Stretch Assays for Studies of Protein-DNA Interactions at the Single-Molecule Level *Aurimas Kopūstas*, Mindaugas Zaremba, *Marijonas Tutkus* Published in Applied Nano, January 2022 (see publication ) - Review, Corresponding author |
 | Oriented Soft DNA Curtains for Single-Molecule Imaging *Aurimas Kopūstas*, *Šarūnė Ivanovaitė*, Tomas Rakickas, Ernesta Pocevičiūtė, *Justė Paksaitė*, Tautvydas Karvelis, Mindaugas Zaremba, Elena Manakova, *and Marijonas Tutkus* Published in Langmuir, March 2021 (see publication ) - Research paper, Corresponding author |
 | Fixed DNA Molecule Arrays for High-Throughput Single DNA–Protein Interaction Studies *Marijonas Tutkus*, Tomas Rakickas, Aurimas Kopu̅stas, Šaru̅nė Ivanovaitė, *Oskaras Venckus*, Vytautas Navikas, Mindaugas Zaremba, Elena Manakova, and Ramu̅nas Valiokas Published in Langmuir, April 2019 (see publication ) - Research paper, first author |
 | Probing the dynamics of restriction endonuclease NgoMIV-DNA interaction by single-molecule FRET *Marijonas Tutkus*, Giedrius Sasnauskas, Danielis Rutkauskas Published in Biopolymers, October 2017 (see publication ) - Research paper, first author |
 | DNA-Endonuclease Complex Dynamics by Simultaneous FRET and Fluorophore Intensity in Evanescent Field *Marijonas Tutkus*, Tomas Marciulionis, Giedrius Sasnauskas, Danielis Rutkauskas Published in Biophysical Journal, March 2017 (see publication ) - Research paper, first author |











