Aim of our research
Our research team works on developing and application of bionanotechnological platforms and cutting-edge fluorescence microscopy techniques to understand life at the molecular scale. Using these techneques we are trying to answer many interesting questions including these:
- How do gene editing tools and other nucleic acid-interacting proteins find their target sites? What determines their specificity? How gene editing affects cell health over several generations?
- Can we find new (and hopefully better) ways to perform gene-editing more safe?
- How can we improve gene-eting tools by looking at single molecules or single cells?
- What are the mechanisms of energy transfer and energy transfer quenching (mainly NPQ) in photosynthesis? How lipid enviroment and antenna complex density affects NPQ process?
Main topics
DNA-protein interaction occurs on a nanometer length scale at millisecond-to-second timescales. To monitor interactions occurring on the seconds time scale surface immobilization of one of interacting partners is often required. These interactions can be visualized at great detail through: force spectroscopy, fluorescence, Forster resonance energy transfer (FRET), atomic force microscopy (AFM), nanopores, DNA flow-stretch assays, etc [A. Kopūstas, et al. Applied Nano, 2022]. In general, single-molecule FRET (smFRET) approach worked very well for many different protein-DNA interaction studies.
Restriction endonucleases (REases) in bacteria and archaea function as defense against invading foreign DNA systems. Over the past 10 years we studied three different REases using smFRET approach: Ecl18kI protein [M. Tutkus, et al. Biophysical J. 2017] , and NgoMIV [M. Tutkus, et al. Biopolymers. 2017]. Since both proteins recognize a palindromic DNA target sequence, DNA fragment in our DNA looping assay could adopt two type of loops (“U”- or “Phi”-shaped) depending on how each copy of protein was oriented on the target site (see figure below this paragraph). In our assay we monitored events of DNA looping that occurred after proteins’ binding onto target sites, and events of DNA loop release that occurred upon proteins dissociation from target sites.
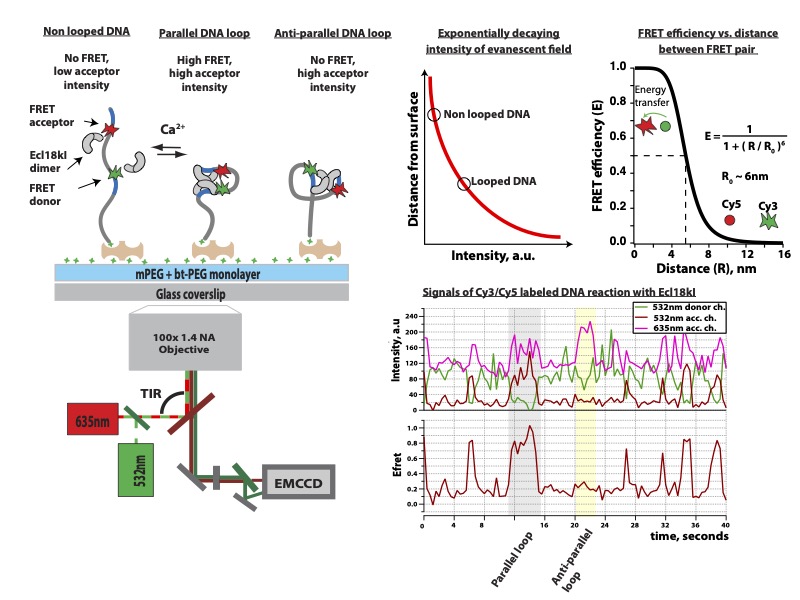
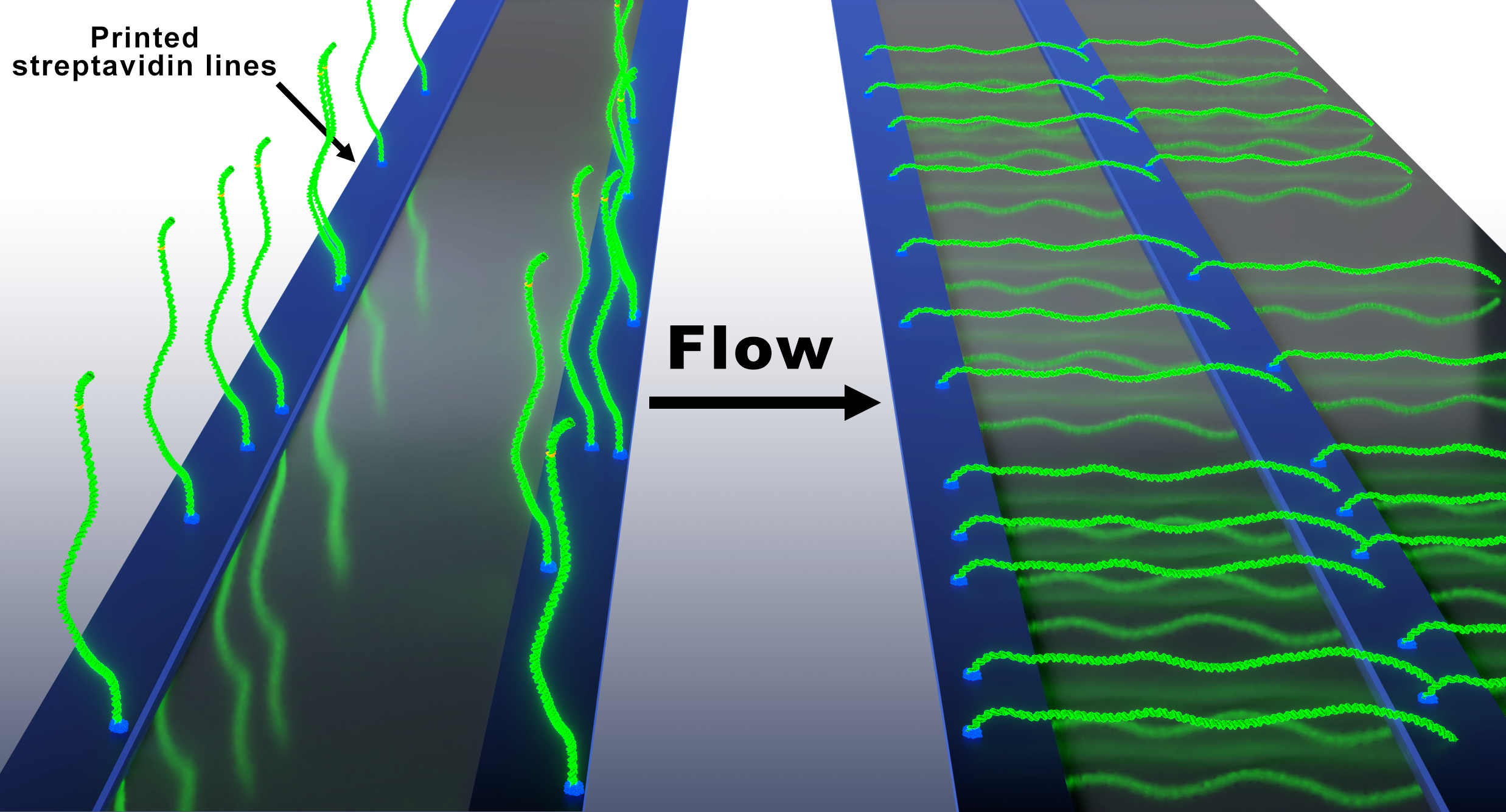
Recently our team developed an alternative assay to the original DNA Curtains that we termed the Oriented Soft DNA Curtains. The main goal of our research is to apply the developed platform for studies of DNA targeting mechanisms of diverse CRISPR-Cas (Clustered regulatory interspaced short palindromic repeats) systems family, novel molecular-tools – prokaryotic Argonaute (pAgo) proteins, and various restriction endonucleases. Since 2017 for these studies we received two grants from the Lithuanian research council. Also, we received funding for a post-doc project dedicated for pAgo studies.
The original DNA Curtains technique is still a little challenging (especially the chromium nanofabrication part), and so naturally we decided to develop a more easy approach. We believe, that the big advantage of our Soft DNA Curtains system is its versatility for single or double tethers. With the same Si-master one can produce both single- and double-tethered Soft DNA Curtains. In case you are interested in the Soft DNA Curtains technology, please contact us using email provided on the left hand side of this page. We are happy to help you with suitable Si-masters, ink, functionalized DNA, and portable semi-automated machines for protein lift-off micro-contact printing.
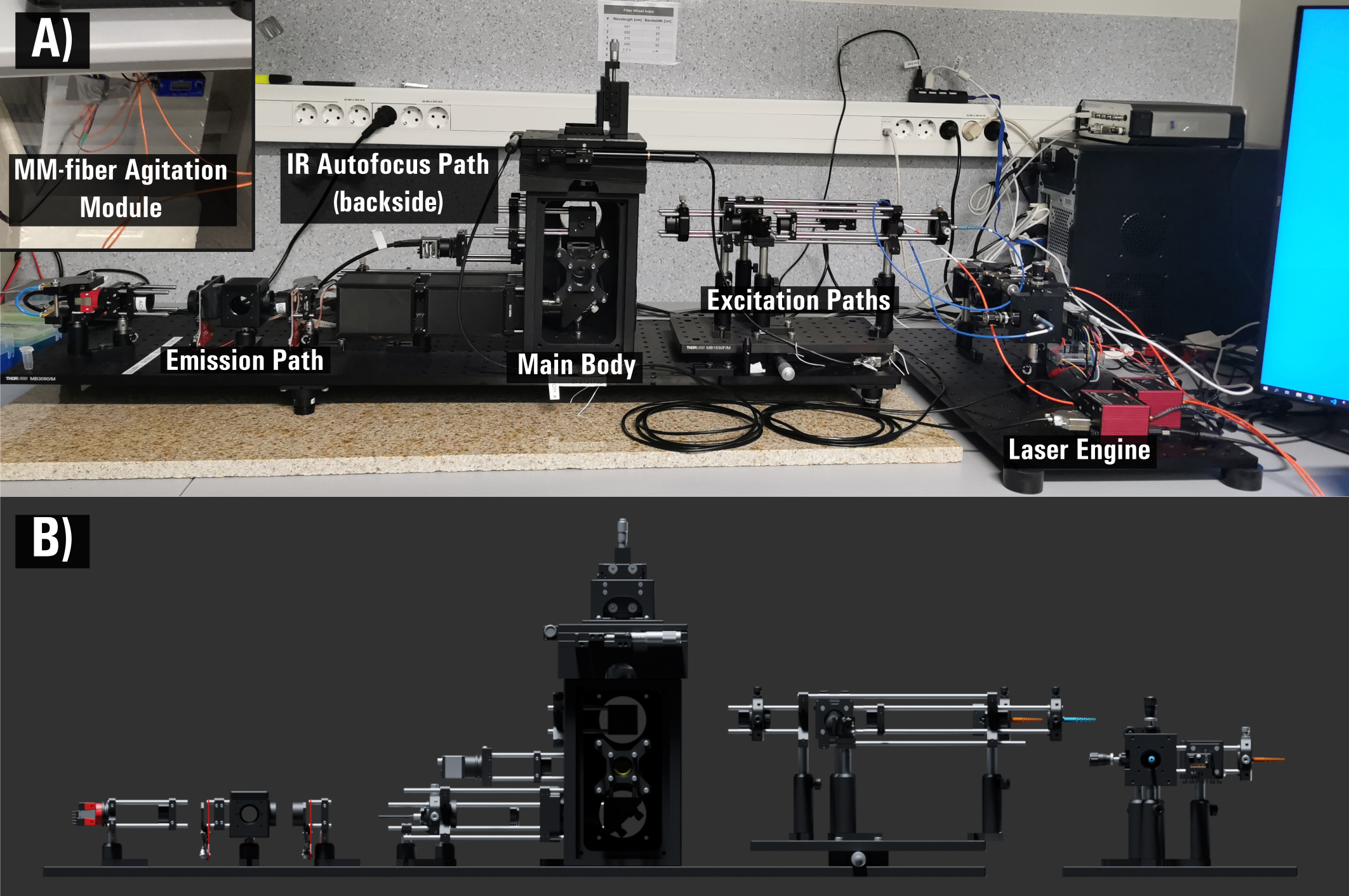
With the help of this LRC funding and our recent grant from Vilnius University Science Fund (MSF-JM-10/2021) we started development of a home-made fluorescence microscopy dedicated for super-resolution studies. This project involves excitation (TIRF and MM-fiber output formation on the imaging plane), detection and autofocusing systems development and industrial-grade CMOS camera application for such studies. Recently our new set-up was cited Nature Methods. Also, we are working on Phyton-based microscope control and single-molecule localization microscopy data analysis (SMLM) packages, and the full discription is available on our github page microEye. The whole aforementioned hardware and software system was recently published as a fully open-source open-microscopy project that we entitled miEye HardwareX. Our miEye microsocpy system was cited in Instruct-LT Annual Report Highlights and New Technologies of the year 2022.
With the help of my recent VU LSC-EMBL Partner Institute Associated Research Fellowship we started studies of chromosomal rearrangements in gene-edited living yeast cells using super-resolution microscopy and fluidics devices in real time. In this project we employ CRISPR-Cas effector proteins - Cas9 (blunt cut) or Cas12a (sticky-end cut) - to introduce DNA double-stranded breaks (DSBs) at different chromosomal locations in the genome of living fission yeast cells. We select targets based on factors including their chromosomal location, 3-D position in the nucleus, and their epigenetic state. Fission yeast cells have similar genome repair, architecture and regulation to humans, but with smaller genomes and single-cell tractability. Cells loaded into individually-addressable positions within the fluidics device, allows for their long-lasting imaging. We track DSBs introduction upon induction of Cas protein expression using existing SR-compatible markers (FROS array) and also develop new ways of labeling specfic genome locus with high singal-to-background ratio. Movement of markers reflects chromosome rearrangement that we can track using SR microscopy. For these studies we employ our developed miEye hardware and software system HardwareX.
The Laboratory
We’re based at the: Life Sciences Center at the Vilnius University
Center for Physical Sciences and Technology FTMC.
